Lanthanum »
PDB 1djg-9ceq »
2i18 »
Lanthanum in PDB 2i18: The Refined Structure of C-Terminal Domain of An Ef-Hand Calcium Binding Protein From Entamoeba Histolytica
Lanthanum Binding Sites:
The binding sites of Lanthanum atom in the The Refined Structure of C-Terminal Domain of An Ef-Hand Calcium Binding Protein From Entamoeba Histolytica
(pdb code 2i18). This binding sites where shown within
5.0 Angstroms radius around Lanthanum atom.
In total only one binding site of Lanthanum was determined in the The Refined Structure of C-Terminal Domain of An Ef-Hand Calcium Binding Protein From Entamoeba Histolytica, PDB code: 2i18:
In total only one binding site of Lanthanum was determined in the The Refined Structure of C-Terminal Domain of An Ef-Hand Calcium Binding Protein From Entamoeba Histolytica, PDB code: 2i18:
Lanthanum binding site 1 out of 1 in 2i18
Go back to
Lanthanum binding site 1 out
of 1 in the The Refined Structure of C-Terminal Domain of An Ef-Hand Calcium Binding Protein From Entamoeba Histolytica
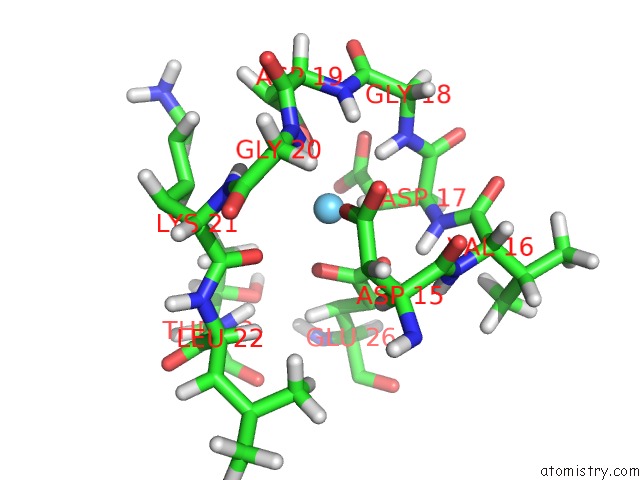
Mono view
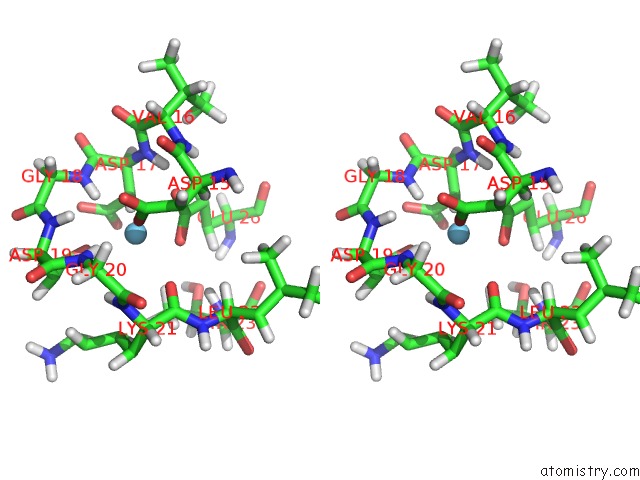
Stereo pair view

Mono view

Stereo pair view
A full contact list of Lanthanum with other atoms in the La binding
site number 1 of The Refined Structure of C-Terminal Domain of An Ef-Hand Calcium Binding Protein From Entamoeba Histolytica within 5.0Å range:
|
Reference:
S.M.Mustafi,
S.Mukherjee,
K.V.Chary,
G.Cavallaro.
Structural Basis For the Observed Differential Magnetic Anisotropic Tensorial Values in Calcium Binding Proteins. Proteins V. 65 656 2006.
ISSN: ISSN 0887-3585
PubMed: 16981203
DOI: 10.1002/PROT.21121
Page generated: Sat Aug 9 19:53:25 2025
ISSN: ISSN 0887-3585
PubMed: 16981203
DOI: 10.1002/PROT.21121
Last articles
Mg in 1SO2Mg in 1SO5
Mg in 1SO4
Mg in 1SO3
Mg in 1SNF
Mg in 1SLH
Mg in 1SL2
Mg in 1SL5
Mg in 1SKR
Mg in 1SL0